
The first dataset are used for (FNN, LVQ and PNN) the second one is used for CNN and CNN fine tuning. We used this dataset from bioinformatics information Lab from University of Missouri, United States of America, it is contains 1854 protein. The second dataset is formed of proteins from four different classes. The first dataset is obtained from matlab math work and from thesis, it is contain 114 protein samples which are divided into training dataset with 75 protein samples and testing dataset with 44 protein sample. In this section, we will introduce dataset description, measures of prediction accuracy. The main objective of this work is to gain an improvement of prediction accuracy (Q 3) so that the implementation results show that the proposed model (CNN Fine Tuning) performs better than the other models and looks promising for problems with characteristics similar to that problem (PSSP) by achieving prediction accuracy with Q 3 = 90.31%. In this research, the authors have proposed five models of NN that has been used, including FNN, LVQ, PNN, CNN and CNN Fine tuning for PSSP. Therefore, PSSP remains as an active area of research, and an integral part of protein analysis. Protein secondary structure is also used in a variety of scientific areas, including proteome and gene annotation. Protein folding, or the prediction of the tertiary structure from linear sequence, is an unsolved and ubiquitous problem that invites research from many fields of study, including computer science, molecular biology, biochemistry and other. PSSP provides a significant first step toward the tertiary structure prediction, as well as offering information about protein activity, relationship, and function.

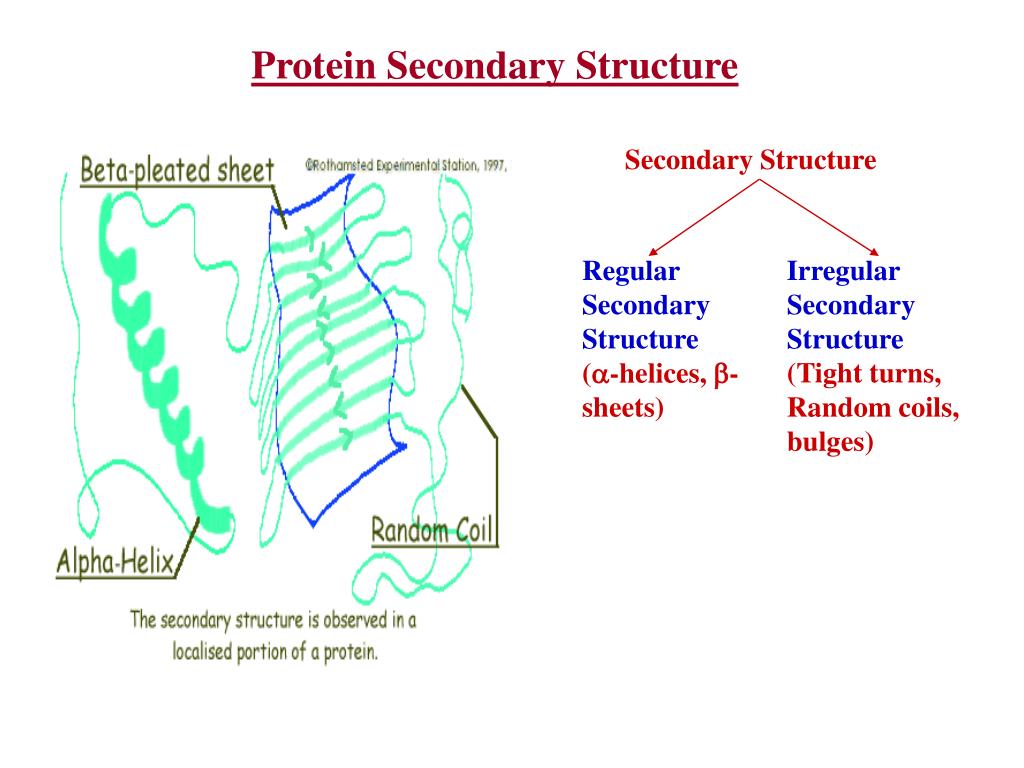
The Secondary structure refers to the arrangement of connections within the amino acid groups to form three different structured classes (H, E, and C).

It provides the foundation of all the other types of structures. The Primary structure contains a sequence of 20 different types of amino acids. There are four different structure types of proteins, namely the Primary, Secondary, Tertiary and Quaternary structures. Protein structure prediction methods are categorized under bioinformatics which is a broad field that combines many other fields and disciplines like biology, biochemistry, information technology, statistics, and mathematics. The use of computers is absolutely essential in mining genomes for information gathering and knowledge building. Bioinformatics involves the technology that uses computers for storage, retrieval, manipulation, and distribution of information related to biological macromolecules such as DNA, RNA, and proteins.


 0 kommentar(er)
0 kommentar(er)
